Fifteen years on the cutting edge of genomics
HudsonAlpha continues driving innovation in genomic research to improve the human condition

For 13 years, an international team of scientists worked tirelessly to sequence the first near-complete human genome. The Human Genome Project changed the course of genetics as we know it, but it wasn’t just about the human genome sequence. The technology and international collaboration born from the initial sequencing project revolutionized the information we can unearth within the genetic code.
The project's success inspired local entrepreneurs Jim Hudson and Lonnie McMillian to create a non-profit research institute focused on using genomics to improve the human condition. With the help of Rick Myers, PhD, former leader of the Stanford University Department of Genetics and the Stanford Human Genome Center, the HudsonAlpha Institute for Biotechnology opened its doors in Huntsville, AL, in 2008. From the beginning, HudsonAlpha aimed to bridge the gap between basic discoveries and real-world applications in biotechnology, continuing the groundbreaking team-based science proven successful by the Human Genome Project.










In 2007, Myers made a daring decision to leave Stanford and return to his home state of Alabama to establish a world-class research team at HudsonAlpha. With a clear mission in mind, Myers handpicked the most talented and skilled scientists from various genetics specialties. HudsonAlpha Faculty Investigators Jane Grimwood, PhD, and Jeremy Schmutz were integral members of the Stanford Human Genome Center team and contributed significantly to the groundbreaking Human Genome Project. They joined Myers at HudsonAlpha in 2008 and began the HudsonAlpha Genome Sequencing Center (GSC) as its co-Directors and Faculty Investigators. The GSC helps drive the institute's cutting-edge research to new heights by providing high-quality genome sequencing and analysis to internal and external collaborators.
“When I first began talking with Jim and Lonnie about HudsonAlpha, I was just serving as an advisor, helping them work through what could be. At some point, when I really stepped back and looked at what they wanted to build, I knew I wanted to be a permanent part of the Institute."
Scientists relied on a DNA sequencing technique called Sanger sequencing to sequence the first human genome. Sanger sequencing mimics the natural process cells use to copy DNA to read the genetic code and piece together small sections of DNA sequence. It offered unparalleled accuracy and was pivotal in the success of the Human Genome Project. When HudsonAlpha began, it boasted 16 Sanger sequencing machines, which were the backbone of genomics research at the time. But the researchers who came to HudsonAlpha had already started to embrace newer, “next-generation” sequencing technologies, anticipating they would soon quickly replace the technologies they used in the Human Genome Project.
The Human Genome Project’s triumph fueled rapid advancements in DNA sequencing technology as scientists were eager to find ways to make the process faster and more affordable. The next wave of genetic sequencing technology supplanted the Sanger sequencing approach and ramped it up dramatically. Like Sanger sequencing, automated next-generation sequencing identifies the bases of a small section of DNA. But instead of doing it for just a few fragments at a time, it sequences millions of fragments simultaneously. Today, next-generation sequencing platforms like Illumina’s NovaSeq and Pacific Bioscience’s Onso can sequence a whole human genome in just one day and for a tiny fraction of the cost of the first one.
For the past 15 years, HudsonAlpha’s research labs committed to staying at the forefront of sequencing technology. As new sequencing technologies emerged, HudsonAlpha’s GSC was fortunate to serve as pilot testers, giving the new machines a run for their sequencing money.
“After Sanger sequencing, our equipment evolved with the ever-changing landscape of technology. We’ve transitioned through several next-generation sequencing platforms over the past 15 years, each pushing the limits of how quickly, efficiently, and cost-effectively we could sequence DNA. Today, we run seven PacBio Sequels and an Illumina NovaSeq 6000, with a brand new PacBio Revio on the horizon.”
Although next-generation sequencing technology is great at spotting small genetic changes, there are better tools for detecting larger genetic rearrangements impacting more than fifty bases, like deletions, duplications, inversions, or translocations. Enter the next wave of sequencing technology: real-time, single-molecule DNA sequencing platforms. Instead of producing millions of short sections of DNA sequences that must be reassembled, these long-read sequencing platforms produce sequence reads that are up to 1,000 times longer.
Long-read sequencing provides many advantages over next-generation sequencing, including producing reads of tens of thousands of bases, enabling scientists to capture more complex regions and detect DNA epigenetic modifications. Although longer reads are expensive, researchers continuously develop and refine technologies to produce highly accurate, long reads at a lower cost, expanding genomic understanding and revolutionizing genetic research.
With each new iteration of sequencing technology came the ability to create more genomic data beyond anything scientists had dealt with before. Now, instead of creating single genomes for each species, scientists can sequence and analyze many genomes, gaining a better picture of genetic diversity.
“The more data we can produce, the more we can learn about plant and animal genomics. With the newest sequencing platforms, we have the highest throughput at the lowest cost we’ve ever seen. Seeing the downstream applications of this almost unending stream of new data and how we’re building knowledge networks within and across species is truly exciting.”




"None of this would have been possible without the investments in technology and the culture of big picture science that Rick, Jim, and others laid the foundation for."

Collaborative Science
The Human Genome Project was one of the most extraordinary collaborative efforts in scientific history, involving thousands of researchers worldwide. It inspired a new era of collaborative science, where scientists from different fields work together to tackle complex problems. HudsonAlpha scientists continue to be integral members of many large consortium projects.
ENCODE
The Encyclopedia of DNA Elements (ENCODE) Consortium began almost immediately after the Human Genome Project and ran for 20 years. Funded by the National Human Research Institute, ENCODE’s goal was to identify all the functional elements in the genome, such as genes, regulatory regions, and non-coding sequences, and decipher their role in gene expression, cell differentiation, and disease development. The Consortium created a comprehensive database of genomic information that is freely available to the scientific community. Scientists at HudsonAlpha, led by Dr. Myers, focused their ENCODE efforts primarily on studying transcription factors that control gene expression. Although the fourth and final chapter of ENCODE ended in 2022, the vast amounts of public data produced through ENCODE will lead to groundbreaking discoveries for years to come.
ReDLat
HudsonAlpha Faculty Investigators Rick Myers, PhD, and Nick Cochran, PhD, are members of the Multi-Partner Consortium to Expand Dementia Research in Latin America (ReDLat). The consortium is a multinational effort to expand dementia research in Latin America, particularly in underserved and diverse populations. The main goal is to identify the genetic and socioeconomic factors that contribute to Alzheimer's disease and other dementias in this region. Over five years, the consortium plans to collect data from over 4,000 individuals across several countries, including Argentina, Brazil, Chile, Colombia, Mexico, Peru, and the US. This comprehensive approach will help to provide a unique understanding of the genetic and environmental underpinnings of dementia in Latin America, ultimately leading to better prevention and treatment strategies for affected individuals.
All of Us
The All of Us research program was launched by the National Institutes of Health (NIH) in 2015 to enroll and collect health data from one million Americans, creating one of the largest, most diverse public resources for biomedical research in human history. All of Us is committed to using cutting-edge techniques to collect and analyze the program’s data. One example of this is using long-read sequencing technology to sequence DNA samples. In 2019, the first set of long-read sequencing data was made possible by scientists at HudsonAlpha in collaboration with Discovery Life Sciences (DLS), a company located on the HudsonAlpha campus. Today, HudsonAlpha is on track to complete a long-read genome for more than 2,000 All of Us participants.
Staying up-to-date with the latest sequencing technologies has paid off for HudsonAlpha. Over the past 15 years, HudsonAlpha researchers made groundbreaking discoveries in human genetics, basic biology, plant genetics, and various human disease fields, including autoimmune disease, neurological and neuropsychiatric diseases, cancer, and rare diseases.

Whole genome sequencing for disease diagnosis
By analyzing an individual's entire genetic code, doctors can uncover the root cause of a patient's symptoms and develop personalized treatment plans. In the decades since the completion of the Human Genome Project, whole genome sequencing technology has advanced and become more cost-effective. In 2011, scientists first used whole genome sequencing to diagnose a patient whose disease had stumped physicians for years. Since then, whole genome sequencing has revolutionized how scientists and physicians diagnose and treat diseases.
HudsonAlpha Faculty Investigator Greg Cooper, PhD, and his lab are experts at using genome sequencing to diagnose rare diseases. Over the past decade, Cooper’s lab and many collaborating labs sequenced the genomes of more than 1,790 children with rare diseases, affording diagnoses to about 27 percent of patients.
Recognizing the importance of genetic sequencing to clinical care, HudsonAlpha founded the Smith Family Clinic for Genomic Medicine, LLC, in 2015. It is a stand-alone medical office residing on HudsonAlpha’s campus that focuses on diagnosing rare genetic diseases using genomics. Cooper’s lab works closely with the clinic to help find genetic explanations for patients’ symptoms.
In addition to helping patients with rare diseases find answers, HudsonAlpha scientists are also part of a collaborative project to promote new discoveries for children affected with cancer and structural birth defects. As part of the Gabriella Miller Kids First Pediatric Research Program (Kids First), HudsonAlpha is one of the sequencing centers for the program. In collaboration with DLS, the center sequenced thousands of samples to date while offering reliable and efficient data storage and access capability that provides access, sharing, and reporting to Kids First research program directors and investigators.
The joint project provides whole genome sequencing and analysis, RNA sequencing, and whole exome sequencing for pediatric cancer samples. During the first seven years of this program, data from approximately 30,000 participants were generated and made available following the sequencing of DNA, and some RNA, samples from pediatric cancer and structural birth defects projects. Ongoing projects aim to increase the number of patient samples sequenced to elucidate genetic contributions to childhood cancers and the etiology of structural birth defects.









Gene Discoveries
The advances in genetic sequencing technology that followed the completion of the Human Genome Project have profoundly impacted our understanding of the genetic basis of diseases. With the ability to sequence the entire human genome more efficiently and accurately than ever, HudsonAlpha scientists identified genetic variations associated with a wide range of diseases, from rare genetic disorders to common complex diseases such as cancer, neurodegenerative diseases, and autoimmune diseases. This has led to a better understanding of the underlying mechanisms of these diseases and has enabled the development of new diagnostic tools and targeted therapies that are more effective and personalized.
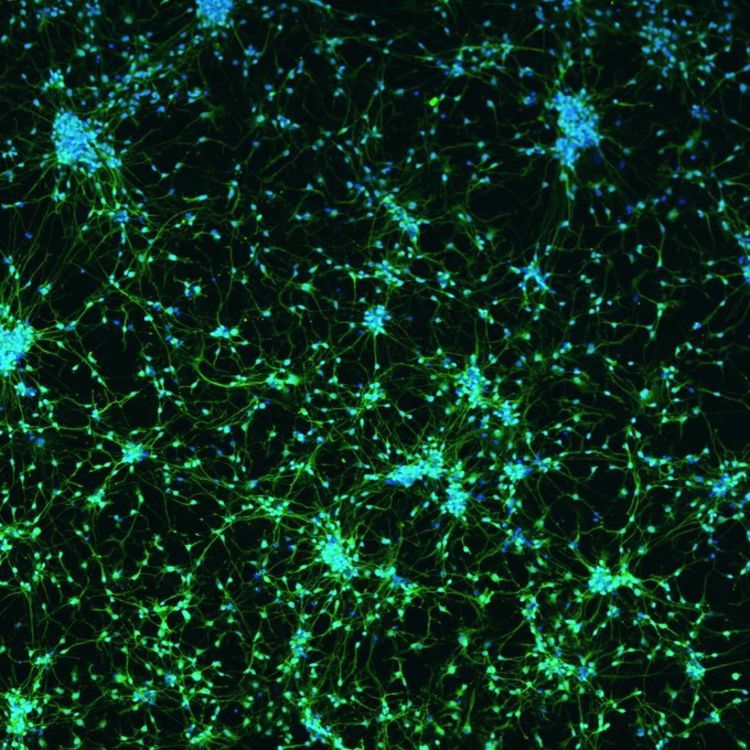
Neurodegenerative diseases
Neurodegenerative diseases are devastating, slowly robbing individuals of their cognitive ability, memory, mobility, and, eventually, their lives. At HudsonAlpha, scientists use genetic technology to identify the genes responsible for these conditions. By studying the DNA of affected individuals, they can pinpoint specific genetic variations more common in people with the disease. But HudsonAlpha goes beyond identification. Scientists also analyze how these genetic variants cause symptoms at the molecular level. Understanding these diseases' underlying biology can help develop targeted therapies and interventions to treat or delay them. Over the past decade, HudsonAlpha researchers Myers and Cochran discovered several genes linked to neurodegenerative diseases, including ALS, Alzheimer's disease, and frontotemporal dementia. This important research brings us closer to a future where we can more effectively treat and ultimately cure these harrowing conditions.
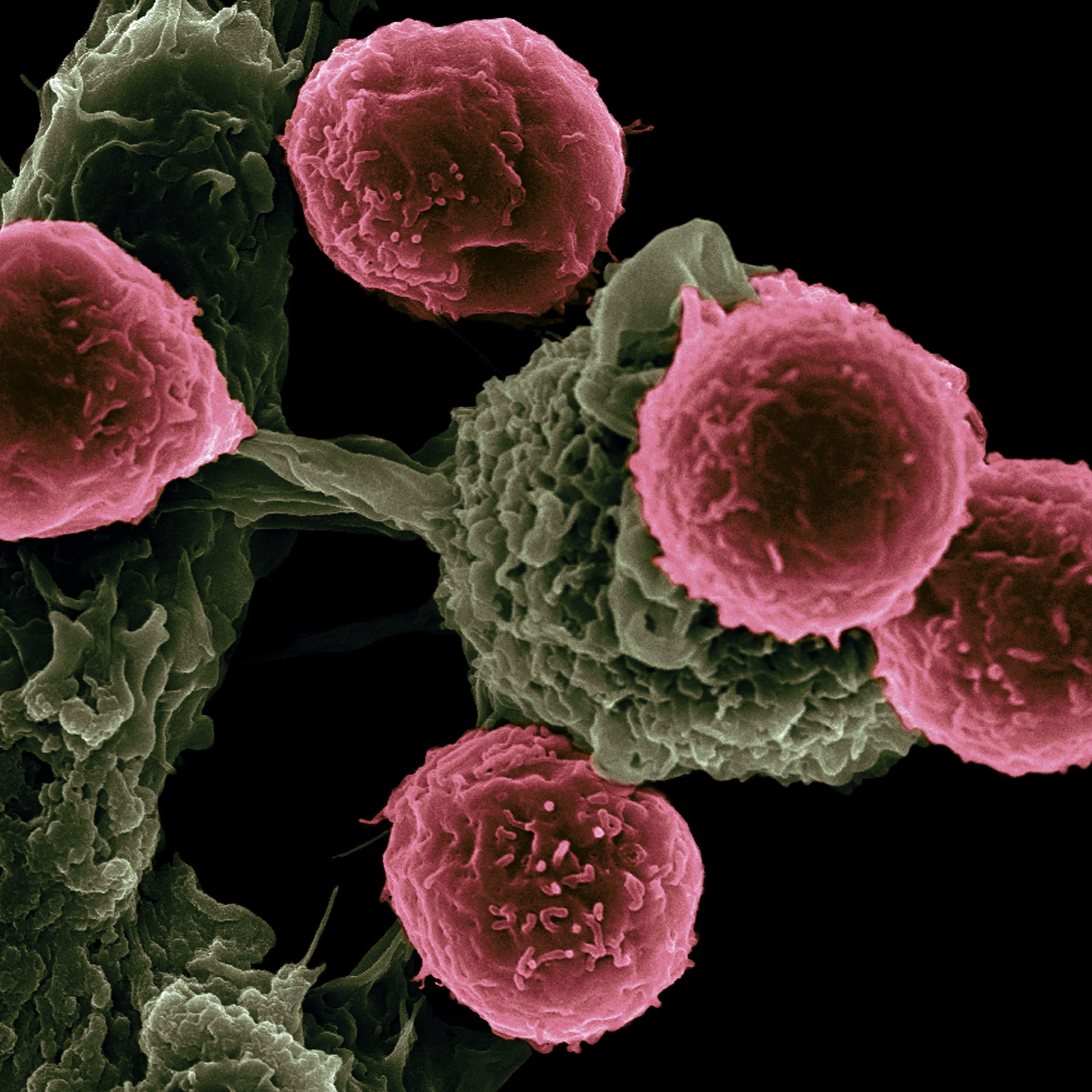
Cancer
Genomic technology is also revolutionizing the field of precision oncology. HudsonAlpha scientists have been at the forefront of this cutting-edge field, using genomic technology to better understand how cancer patients respond to treatment. By analyzing the DNA of cancer cells, they can identify specific genetic variations that may impact how a patient responds to a particular therapy. This knowledge is critical for predicting treatment outcomes and guiding physicians to use alternative treatments when needed. The groundbreaking work of HudsonAlpha scientists, including faculty investigators Sara Cooper, PhD, and Myers, has led to important findings in breast, ovarian, pancreatic, prostate, and colon cancers. With continued advances in genetic technology, we are moving closer to a future where cancer treatment is personalized and more effective than ever before.

Neurodevelopmental diseases
Two out of every 100 children are born with a physical disability or developmental delay, which often arises from genetic factors. Dr. Greg Cooper's lab is a global leader in identifying new rare disease variants that can help diagnose and treat these conditions. By connecting with other researchers who have found variants in the same gene, Cooper's team has confirmed more than a dozen genes likely to cause developmental disorders, such as EBF3, RALA, BRSK2, and ZMYM3. Their collaborative efforts have resulted in 25 publications and are paving the way for better understanding and treatment of rare diseases. Cooper’s lab recently began using long-read sequencing technology to help physicians make diagnoses for pediatric patients affected by undiagnosed neurodevelopmental disorders who received no definitive diagnosis using other sequencing technology.
“The way in which we think about human biology and disease is facilitated by advances in technology like DNA sequencing but rests on a basic understanding of how genes and cells interact. Just as Greger Mendel discovered genes by studying the simple pea plant, geneticists often use simpler systems to uncover principles of developmental biology that apply to all animals, including humans.”
-Greg Barsh, MD, PhD, Faculty Investigator and Smith Family Chair in Genomics

HudsonAlpha Faculty Investigator Greg Barsh, MD, PhD, is a medical geneticist who studies the genes and mechanisms that underlie pattern formation during mammalian development. Fingers, toes, blood vessels, and airways in the lung all depend on pattern formation during development. Barsh studies a different system offering unique advantages: color patterns in domestic and wild mammals. Over the last 15 years, his work on “How the leopard gets its spots” has led to fundamental new insight into developmental mechanisms that act across different tissues in all mammals, including humans.
In 2012, Barsh and his lab identified the first gene responsible for creating the distinctive patterns of a cat’s coat. The gene, Transmembrane aminopeptidase Q (Taqpep), creates narrow stripe patterns in tabby cats when functioning properly and a blotchy coat pattern if the gene is turned off. In further support of their discovery, they also found that mutations in Taqpep cause standard spots to turn into blotches and stripes in the rare king cheetah. The team determined at least two other genes, including Endothelin 3 (Edn3), produce proteins central to a cascade of cell-level events that generate cats’ distinctive coat colors and patterns.
Although Barsh’s team identified important players in coat color and pattern establishment, when, where, and how the patterns arise were still largely unknown until 2021. Through partnerships with several feral cat trap-neuter-release programs, Barsh’s team studied developing cat skin from fetal tissue that was otherwise discarded during spay procedures. By analyzing the skin microscopically and on a single-cell RNA level, they discovered a gene, Dickkopf 4 (Dkk4), that codes for a signaling molecule that controls color pattern formation. Dkk4 marks areas of fetal skin that give rise to hair follicles that later produce dark pigment. Various mutations in Dkk4 are responsible for different cat coat patterns.
The decade-long research project highlights HudsonAlpha researchers’ dedication to staying at the forefront of genetics by using the most cutting-edge techniques and embracing collaborative research practices.
Tackling the Complex
The availability of high-throughput sequencing platforms, innovative bioinformatics tools, and the development of long-read sequencing technologies have made it possible to sequence even the most complicated plant species with high accuracy and completeness. This allows researchers at HudsonAlpha to study the genetic basis of plant traits, such as disease resistance, drought tolerance, and yield, and to identify key genes and pathways involved in these processes.

A ten-year reference genome
Through one decade-long sequencing project, HudsonAlpha scientists experienced in real-time how advances in sequencing technology improved the quality and ease of assembly of complicated genomes. Switchgrass, a native North American grass with a wide range spanning from Canada to southern Mexico, is a promising candidate for bioenergy production. The first switchgrass reference genome, sequenced in 2008, was created using Roche 454 sequencing, an early transition technology from Sanger.
Over the next decade, GSC scientists added parts to that genome using each new sequencing technology. A high-quality reference genome was finally achieved using long-read sequencing information in 2020. The publicly available reference genome enabled collaborators to compare the genomes of two types of switchgrass and unlock important insights into its genetic makeup. The genetic tools created by HudsonAlpha’s expert sequencing team are helping optimize switchgrass to grow efficiently in diverse environments, ensuring its success as a sustainable, eco-friendly energy source for the future.

Towards pangenomes
Pangenomes, the collective genome of a species, are revolutionizing the field of genomics by providing a more comprehensive understanding of genetic variation and evolution within a species. Unlike traditional reference genomes, which are derived from a single individual and represent a limited set of genetic variation, pangenomes incorporate genetic diversity from multiple individuals or strains, allowing for a more complete picture of genomic variation within a species.
In plant genomics, understanding DNA variants that plants need to produce useful traits, such as increased yield and ability to survive extreme weather, can help crop breeders create optimized varieties of crop plants for our changing planet. At HudsonAlpha, plant scientists have embraced pangenomes' importance and usefulness in their research. By looking at many genomes for a species, scientists have recently made discoveries in plants such as switchgrass, green millet, pecan, and barley that would not have been possible without the pangenome.
Founded on the principles established by the Human Genome Project – team science, advancing DNA sequencing technology, and a pure curiosity for the genomic world – HudsonAlpha has continued to grow and evolve those principles to bridge the gap between foundational discoveries and real-world applications. Its unwavering dedication to advancing genomic science and commitment to translating discoveries into tangible solutions make HudsonAlpha a shining example of what is possible when brilliant minds and cutting-edge technology push the boundaries of what we know about the world around us. Stay tuned for the next installment of this series: The Future of Research and Innovation at HudsonAlpha.










Images: Adobe Stock, Unsplash, HudsonAlpha Archives, Jill Amey, Cathleen Shaw, Kaitlyn Williams, Robert Goodwin